Abstract
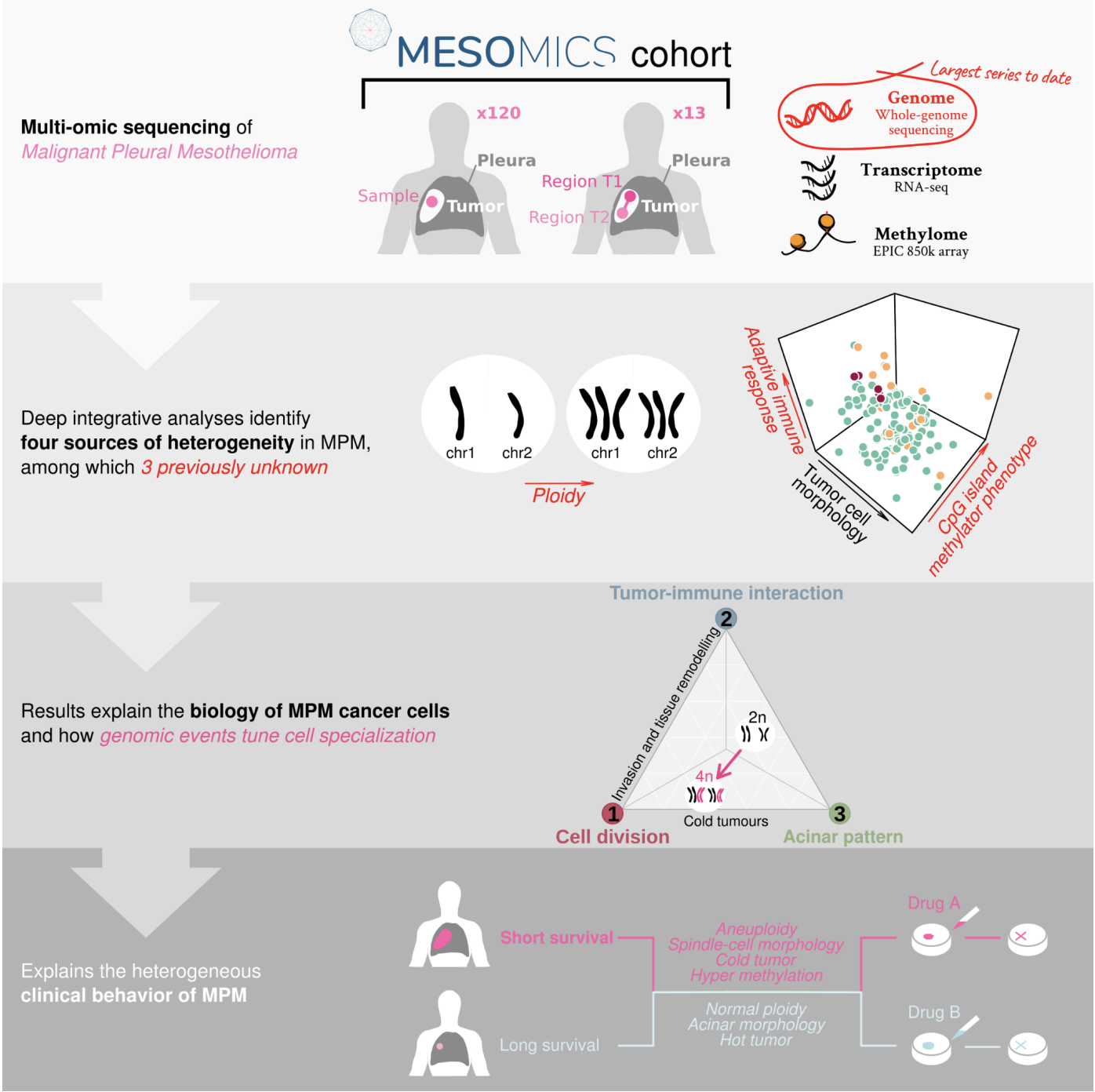
Malignant Pleural Mesothelioma (MPM) is an aggressive cancer with rising incidence and challenging clinical management. Using the largest series of whole-genome sequencing data integrated with transcriptomic and epigenomic data using multi-omic factor analysis, we demonstrate that MPM heterogeneity arises from four sources of variation: tumor cell morphology, ploidy, adaptive immune response, and CpG island methylator phenotype. Previous genomic studies focused on describing only the tumor cell morphology factor, although we robustly find the three other sources in all publicly available cohorts. We prove how these sources of variation explain the biological functions performed by the cancer cells, and how genomic events shape MPM molecular profiles. We show how these new sources of variation help understand the heterogeneity of the clinical behavior of MPM and drug responses measured in cell lines. These findings unearth the interplay between MPM functional biology and its genomic history, and ultimately, inform classification, prognostication and treatment.
Press Coverage
- Lifting the curtain on molecular differences between malignant pleural mesotheliomas, Nature Genetics, Research Briefing (16 March 2023).