Abstract
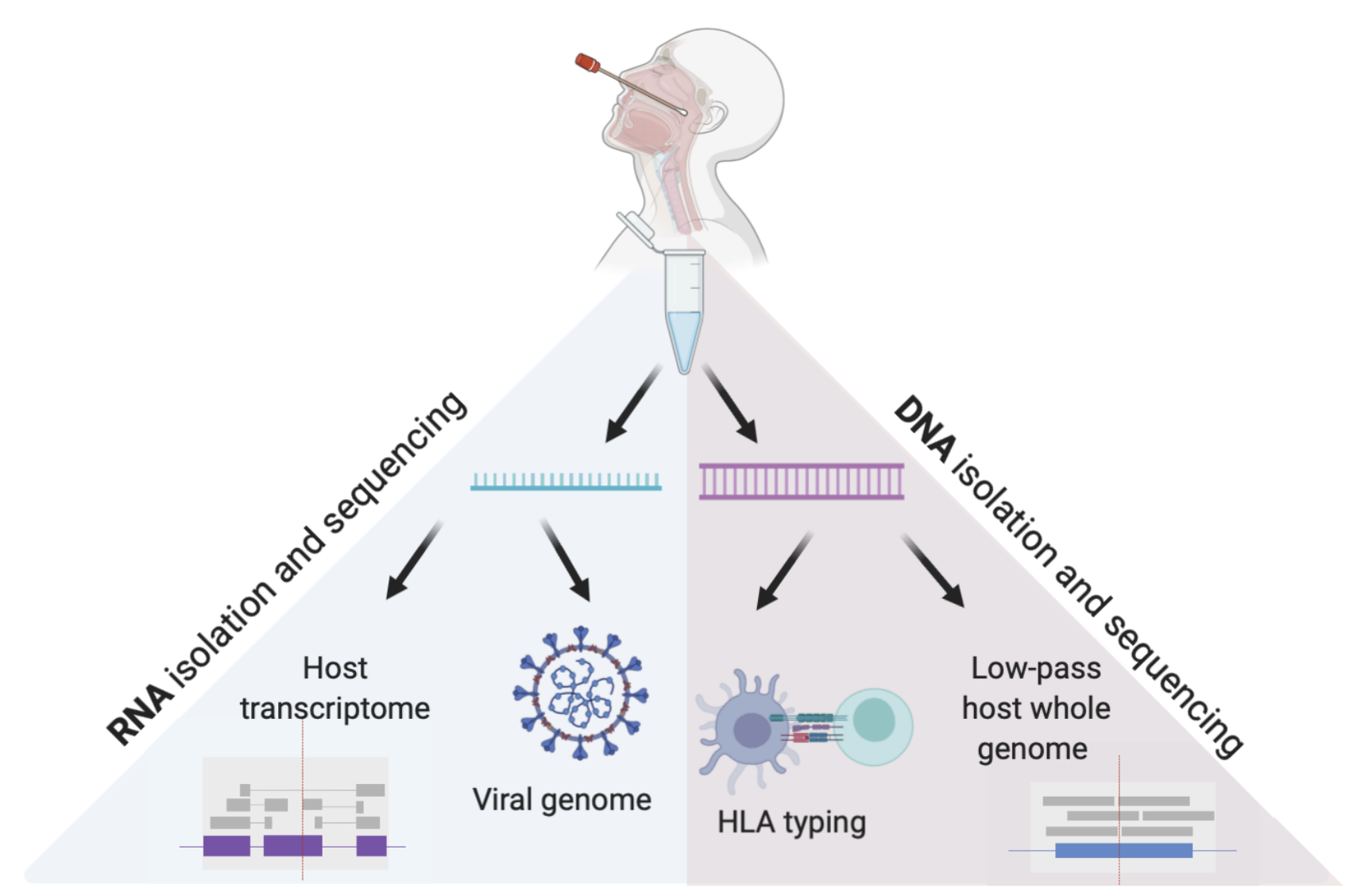
During COVID19 and other viral pandemics, rapid generation of host and pathogen genomic data is critical to tracking infection and informing therapies. There is an urgent need for efficient approaches to this data generation at scale. We have developed a scalable, high throughput approach to generate high fidelity low pass whole genome and HLA sequencing, viral genomes, and representation of human transcriptome from single nasopharyngeal swabs of COVID19 patients.