Abstract
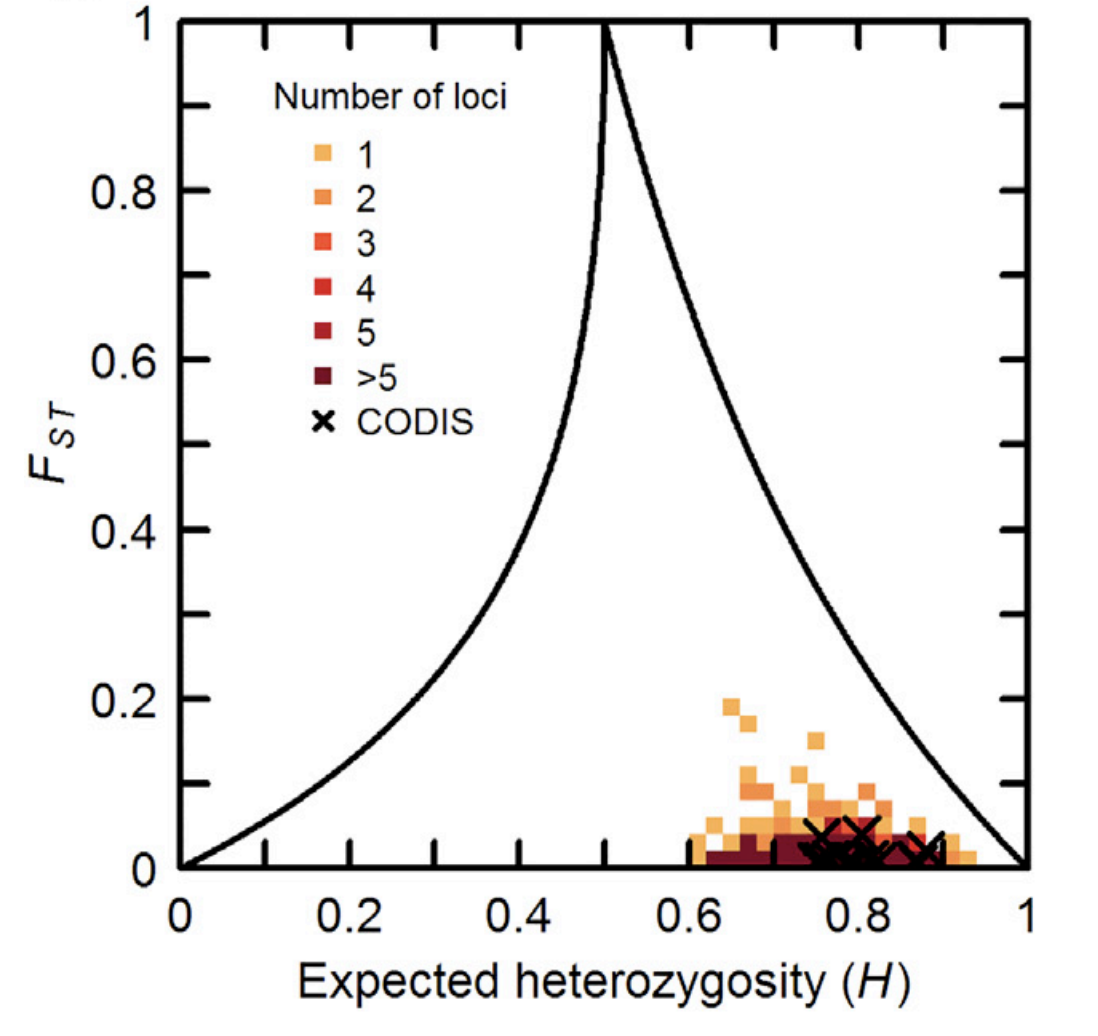
Highly polymorphic genetic markers with significant potential for distinguishing individual identity are used as a standard tool in forensic testing. At the same time, population-genetic studies have suggested that genetically diverse markers with high individual identifiability also confer information about genetic ancestry. The dual influence of polymorphism levels on ancestry inference and forensic desirability suggests that forensically useful marker sets with high levels of individual identifiability might also possess substantial ancestry information. We study a standard forensic marker set—the 13 CODIS loci used in the United States and elsewhere—together with 779 additional microsatellites, using direct population structure inference to test whether markers with substantial individual identifiability also produce considerable information about ancestry. Despite having been selected for individual identification and not for ancestry inference, the CODIS markers generate nontrivial model-based clustering patterns similar to those of other sets of 13 tetranucleotide microsatellites. Although the CODIS markers have relatively low values of the FST divergence statistic, their high heterozygosities produce greater ancestry inference potential than is possessed by less heterozygous marker sets. More generally, we observe that marker sets with greater individual identifiability also tend toward greater population identifiability. We conclude that population identifiability regularly follows as a byproduct of the use of highly polymorphic forensic markers. Our findings have implications for the design of new forensic marker sets and for evaluations of the extent to which individual characteristics beyond identification might be predicted from current and future forensic data.